Rare Variant Analysis and Interpretation
Comprehensive variant analysis and interpretation for rare disease research
Streamlined genetic disease workflows that power labs for growth
Streamlined analysis of genetic variants
Current tools for rare-genetic and other germline variant analysis are fragmented, often involving several pipelines requiring separate integration efforts and maintenance schedules. This disjointed framework prevents laboratories from scaling services effectively. Today, high-throughput labs need intuitive tools for variant calling, insights generation and interpretation support, and reporting in a comprehensive and seamless way.
Emedgene, integrated with DRAGEN, consolidates all these functions at your fingertips. Simplify rare variant analysis and interpretation for research applications by calling, prioritizing, and reporting on variants from a single software interface within a connected workflow. Emedgene is ideal for mid to high-throughput labs using whole-genome, whole-exome, and targeted or virtual panel sequencing to evaluate variants and generate insights associated with genetic disease.
Learn more about DRAGEN Secondary Analysis to support comprehensive, accurate, and efficient variant calling.
Learn more about Emedgene to integrate with DRAGEN and to streamline variant interpretation through research report generation.

Rare variant analysis, streamlined.
Find critical answers quickly with comprehensive variant calling and interpretation.
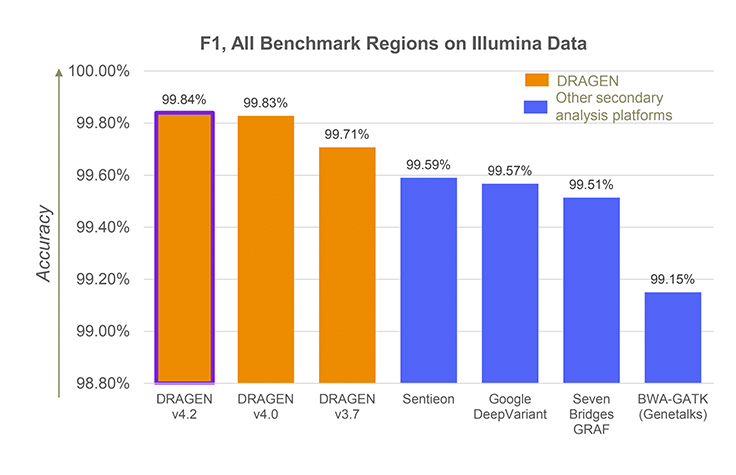
DRAGEN accuracy graph
Most accurate secondary analysis in all-benchmark regions, as compared against all participating solutions in F1 score using PrecisionFDA v2 Truth Challenge Benchmark Data
Accurate, comprehensive variant calling
DRAGEN secondary analysis provides highly accurate rare variant calling and analysis, with automated processing and rapid turnaround time. Powered by the DRAGEN multigenome (graph) reference and machine learning (ML), the suite performs accurate, comprehensive, and efficient identification for key variant types and capabilities, including:
- Small variants
- Copy number variants
- Structural variants
- Mitochondrial variants
- Repeat expansions
- Runs of homozygosity
- Paralogs (eg, SMN1/SMN2)
- Challenging clinically-relevant genes
- Numerous pharmacogenes
- Sex chromosome low allele frequency variants
- Imputation for haploid species and sex chromosomes
By analyzing and visualizing multiple types at one time, you can easily view compound heterozygosity across variant categories. DRAGEN v4.2 functionalities are now available on Emedgene for integrated rare variant analysis and interpretation.

Rare variant analysis and interpretation
Streamline tertiary analysis workflows
Emedgene was designed by geneticists to streamline interpretation workflows and reduce the variant curation burden. It features an automated, explainable-AI (XAI)–powered genomic analysis platform that enables high-throughput interpretation with a 2–5x increase in efficiency and 50–75% reduction in total workflow time per subject.
Explainable AI (XAI) and automation
Never a black box. Emedgene’s XAI prioritizes putative causative variants, backed by transparent evidence, to increase workflow efficiency and confidence. In a study with Baylor Genetics*, Emedgene’s XAI was shown to be 97% accurate (98% for trios) in prioritizing relevant insights. Highly accurate and transparent, XAI can automatically suggest variants for further review in complex data sets that would typically require hours of manual research.

"Emedgene’s machine learning simplifies the highly complex task of variant analysis, allowing us to handle more tests every day."
Dr. Ray Louie, PhD
Assistant Director, Greenwood Genetic Center
Integrated rare variant analysis workflow
By integrating with secondary analysis solutions such as DRAGEN™, data storage platforms such as Illumina Connected Analytics, and electronic health record (EHR) platforms, Emedgene offers a seamless and secure solution from sequencing through research report generation and beyond.
Powered for Growth
Emedgene scales WGS, WES, targeted panels, and virtual panels for variant interpretation across applications and variant types: single nucleotide variants (SNVs), insertions/deletions (indels), copy number variants (CNVs), mitochondrial DNA variants (mtDNA), structural variants (SVs), and short tandem repeats (STRs).
Featured Rare Variant Analysis Articles
Scaling from Exome to Whole-Genome Sequencing with DRAGEN
DRAGEN secondary analysis enables GeneDx to scale to whole-genome analysis and identify variants with precision.
Read ArticleDriving Standards and Best Practices in Clinical Sequencing
The Medical Genome Initiative consortium aims to expand access to high-quality clinical whole-genome sequencing for rare genetic diseases.
Read ArticleStudy Assesses Newborns with Suspected Rare Diseases
A randomized clinical trial examines the effect of whole-genome sequencing on the clinical management of acutely ill infants with suspected genetic disease.
Read ArticleRelated Research Methods
Rare Disease Whole-Genome Sequencing
Whole-genome sequencing is the most comprehensive test for rare disease, with the potential with the potential to uncover new breakthroughs in disease causality.
Learn MoreWhole Exome Sequencing
Whole Exome Sequencing is used to investigate protein-coding regions of the genome to uncover genetic influences on disease and population health.
Learn MoreGenetic and Rare Diseases
Our technology is helping to drive breakthroughs in genetic disease research by facilitating faster insights.
Learn MoreRelated Solutions
Illumina DNA PCR-Free Prep
High performance for sensitive applications such as human whole-genome sequencing.

Illumina sequencing platforms
Get scalable throughput and flexibility for virtually any sequencing method, genome, and scale of project.

Illumina DRAGEN Secondary Analysis
Perform accurate, comprehensive, and efficient analysis of next-generation sequencing data.
Reference
- Miller NA, Farrow EG, Gibson M, et al.A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. doi: 10.1186/s13073-015-0221-8.