
Order the NovaSeq X Series
Advanced chemistry, optics, and informatics combine to deliver exceptional speed and data quality, outstanding throughput and scalability.
This high-throughput NGS assay enables labs to detect SARS-CoV-2 mutations to identify and track the emergence and prevalence of novel variants.
This product is available for Research Use Only (RUO). An in vitro diagnostic version of this product is available for use in Japan, Philippines, and the US under each country's respective authorizing agency.
COVIDSeq Test (RUO Version) is a next-generation sequencing (NGS) assay that enables researchers to detect and characterize novel variants of SARS-CoV-2.
The Illumina COVIDSeq Test (RUO version) accommodates 384 to 3072 samples, depending on throughput needs.
Coverage is particularly focused on the spike protein locus. For detailed sequencing, the optional ARTIC v4 primer pool provides in-depth characterization of new variants.
The kit includes all reagents needed for cDNA conversion, amplification, and library prep. A 63°C annealing temperature during PCR improves variant analysis and insights.
Assess lineage and annotate mutations with open software tools, including the Illumina DRAGEN COVID Lineage App in BaseSpace Sequence Hub.
| Automation capability | Liquid Handling Robots |
|---|---|
| Automation details | Explore available automation methods |
| Content specifications |
Uniform viral genome coverage across the spike protein locus |
| Description |
The COVIDSeq Test (RUO Version) is a scalable high-throughput NGS assay (up to 3072 samples) intended for research applications. |
| Instruments | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode, NovaSeq 6000Dx in Research Mode, NextSeq 500 System, NovaSeq 6000 System |
| Method | Targeted RNA Sequencing, Amplicon Sequencing |
| Multiplexing |
384-plex on NextSeq 384-plex on NovaSeq |
| Nucleic acid type | RNA |
| Sample type details |
Nasopharyngeal, oropharyngeal, and mid-turbinate nasal swab samples |
| Species category | Human, Virus |
| Species details | SARS-CoV-2 (SARS-CoV-2_ MN908947v3) |
| Strand specificity | Non-Stranded |
| Technology | Sequencing |
In addition to the Illumina COVIDSeq Test (3072 samples), you will need:
IDT for Illumina PCR Indexes Sets 1–4 (384 Indexes, 384 samples)
Flow cell for your sequencing system
Sequencing reagent kit for your sequencing system
Viral RNA extraction kit (eg, QIAamp Viral RNA Mini Kit)
The COVIDSeq Test (RUO Version) is an integrated NGS research solution for detection and characterization of SARS-CoV-2, not for use in diagnostic procedures or patient management.
Illumina COVIDSeq Test (RUO Version)
NGS can identify novel coronavirus variants, track COVID-19 transmission, and more. Compare NGS methods for various coronavirus sequencing goals.
As a hypothesis-free method, NGS can distinguish between infectious disease strains that differ by as little as one SNP, and replace the need for multiple tests.
Host genetics and immune response profiling
Studying host genetic differences and individual immune responses to the SARS-CoV-2 virus can shed light on COVID-19 disease susceptibility and severity.
| COVIDSeq Test (RUO Version) | COVIDSeq Assay (96 samples) | Pan-Coronavirus Panel | Respiratory Pathogen ID/AMR Enrichment Panel Kit | |
|---|---|---|---|---|
| Automation capability | Liquid Handling Robots | Liquid Handling Robots | Liquid Handling Robots | |
| Automation details | Explore available automation methods | Explore available automation methods | Explore available automation methods | |
| Content specifications |
Uniform viral genome coverage across the spike protein locus |
Uniform viral genome coverage across the spike protein locus |
Detects respiratory pathogen DNA and RNA simultaneously and profiles antimicrobial resistance (AMR) gene expression concurrently. | |
| Description |
The COVIDSeq Test (RUO Version) is a scalable high-throughput NGS assay (up to 3072 samples) intended for research applications. |
The COVIDSeq Assay (96 samples) is a low- to mid-throughput NGS assay intended to be processed on any Illumina benchtop sequencing system for research applications. | The Pan-Coronavirus Panel is part of an integrated workflow that allows for the detection and whole-genome sequencing of over 200 known and novel coronavirus strains in various animal hosts. | Identify respiratory infections and co-infections, detect antimicrobial resistance markers, and perform strain typing of critical pathogens (SARS-CoV-2 and Flu A/B viruses) to study viral evolution and transmission. |
| Instruments | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode, NovaSeq 6000Dx in Research Mode, NextSeq 500 System, NovaSeq 6000 System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode, NovaSeq 6000Dx in Research Mode, NextSeq 500 System, NovaSeq 6000 System | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System | |
| Method | Targeted RNA Sequencing, Amplicon Sequencing | Targeted RNA Sequencing, Amplicon Sequencing | Target Enrichment, Whole-Genome Sequencing | |
| Multiplexing |
384-plex on NextSeq 384-plex on NovaSeq |
8-plex on the iSeq System 30–48 plex on the MiSeq System 48-plex on the MiniSeq System |
Up to 384 samples in a single run with unique dual indexes | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | RNA | RNA | RNA | |
| Sample type details |
Nasopharyngeal, oropharyngeal, and mid-turbinate nasal swab samples |
Nasopharyngeal, oropharyngeal, and mid-turbinate nasal swab samples |
||
| Species category | Human, Virus | Human, Virus | Virus | |
| Species details | SARS-CoV-2 (SARS-CoV-2_ MN908947v3) | SARS-CoV-2 (SARS-CoV-2_ MN908947v3) | Detects respiratory pathogens (180+ bacteria, 50+ fungi, and 40+ viruses, including SARS-CoV-2) and antimicrobial resistance alleles (1200+). | |
| Strand specificity | Non-Stranded | Non-Stranded | Non-Stranded | |
| Technology | Sequencing | Sequencing | Sequencing |
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
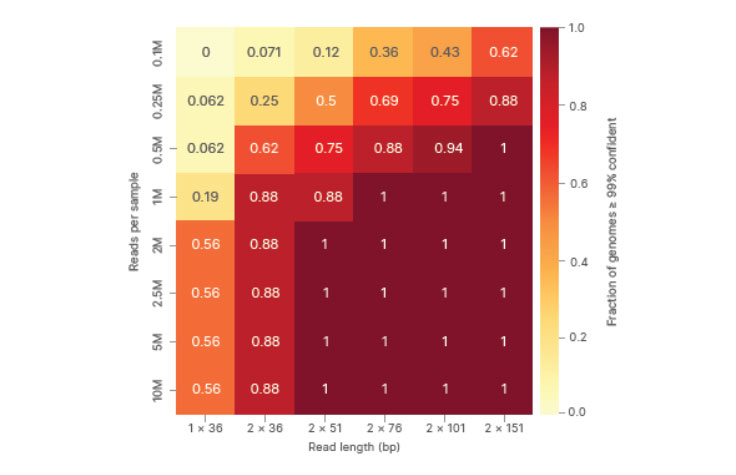

Heat map plotting performance as a function of read length and depth shows significant improvement with paired-end reads over single reads. The fraction of genomes > 0.99 confident is shown, with values close to 1 representing more complete coverage of the SARS-CoV-2 genome.
The COVIDSeq Test (RUO Version) is intended for the NovaSeq 6000 Sequencing System. It can also be run on the iSeq 100, MiniSeq, MiSeq, MiSeqDx in Research Mode, NextSeq 500, NextSeq 550, NextSeq 550Dx in Research Mode, NextSeq 1000, or NextSeq 2000 Systems.
The primary difference is the number of samples each assay can run. The COVIDSeq Assay (96 samples) runs 96 samples on Illumina benchtop sequencing systems to accommodate smaller research labs. In contrast, the COVIDSeq Test (RUO) runs up to 3072 samples on the NovaSeq 6000 System or NextSeq line of sequencing systems.
Yes, an in vitro diagnostic (IVD) test is available for use in authorized countries (Japan, Philippines, and the US) under each country’s respective authorizing agency.
The ARTIC v3 primer pool is included with the COVIDSeq Test (RUO) kit. It contains primers for 98 amplicons and 11 human genes, designed against the original strain of SARS-CoV-2. The optional ARTIC v4 pool includes 99 amplicons and no human controls, designed against the Delta variant. It delivers improved genomic coverage across the spike protein locus and better analytical sensitivity for SARS-CoV-2 variant detection. Read technical note.
Interested in bringing the Illumina COVIDSeq Test into your lab for surveillance applications?
Your email address is never shared with third parties.