Comprehensive analysis of biological systems using environmental DNA
Introduction
Dr. Toshifumi Minamoto,
Genetic Testing Department,
Graduate School of Human Development and Environment, Kobe University
Dr. Toshifumi Minamoto, Graduate School of Human Development and Environment, Kobe University, is a pioneer in environmental DNA research using environmental DNA analysis. He is also committed to establishing an academic society and creating a manual for environmental DNA analysis from water samples. Illumina spoke to Dr. Minamoto about the methodology and significance of environmental DNA analysis, and how it can be applied in multiple research fields in the future.
Q. First, can you explain "environmental DNA" and "environmental DNA analysis"?
A. Environmental DNA analysis is “a specific or comprehensive analysis method” for environmental DNA, which is “DNA released by organisms into the environment”.
The definition of "environmental DNA" has not been well established, even overseas. The definition varies depending on the researcher. In some cases, it may refer to all DNA, such as DNA derived from a living organism, its secretions, or carcasses (or dead microorganisms), including DNA from microorganisms to that of DNA from large organisms. In other cases, it may also only refer to DNA released from the body of an organism, excluding any DNA in the body. I think of it as "a generic term for biological DNA released into the environment."
There are two main directions of the research. One is a species-specific environmental DNA analysis (environmental DNA barcoding) to determine whether specific species, such as fish, are present in the environment and whether the number of that species is large or small. The other is a comprehensive environmental DNA analysis (environmental DNA metabarcoding) to examine species in an entire class, such as fish.
Species-specific environmental DNA analysis is performed mainly by real-time PCR. This requires the design and verification of primers that specifically detect the DNA of target species. On the other hand, comprehensive environmental DNA analysis for fish can be performed by next-generation sequencing (NGS) using universal primers. However, the cost per analysis is higher than species-specific analysis, so it is necessary to take measures, such as collecting a sufficient number of samples before performing the analysis.
Both species-specific and comprehensive analyses are performed depending on the environmental DNA research.
For example, the Opisthorchis viverrini that we are studying first parasitizes shellfish, the infected shellfish are then eaten by the fish which are mainly of the Cyprinidae family and finally the fish are consumed by human. In order to track the route of infection, you will need three kinds of information: that of the parasites, of the shellfish and of the fish. Parasites and shellfish will be subjected to species-specific analysis, and fish will be subjected to comprehensive analysis since various types of fish are parasitized. By accumulating this information, we believe that it is possible to identify where there is a risk of human transmission.
In a survey of Acheilognathus typus, an endangered fish species that is believed to be present in only about 10 locations in four prefectures in the Tohoku region, water samples from 99 locations in Omono River, Akita Prefecture, which is the original habitat of the species, were subjected to both the comprehensive analysis by NGS and the species-specific analysis by PCR. The species was detected only by the species-specific analysis. In fact, Acheilognathus typus was found at a site where a positive result was obtained. Thus, it would be effective to basically use species-specific analysis for targeting rare organisms, and comprehensive analysis when you want to know what kind of relationship that organism has with other organisms.
Q. Please let us know how the eDNA Society created the "Environmental DNA Sampling and Experiment Manual".
A. We established the eDNA Society and created the manual in anticipation of the future accumulation and application of findings.
Five to six years ago, many researchers began to realize that environmental DNA analysis could be used in a variety of research fields. However, there was a concern that if everyone conducted research individually, different methods would be used, which would make accumulating and applying findings difficult. We knew of such cases already happening, at least overseas. At that time, many researchers in Japan were using methods originally developed by our laboratory or methods derived from those we developed. We realized that unification was still possible, so we started to establish the eDNA Society and began preparing the manual. It was great that the first and current chairman of the society, Dr. Michio Kondo (Graduate School of Life Sciences, Tohoku University) took a leading role. The manual was written mainly by members of the eDNA Methods Standardization Committee, and was first published in April 2019.
Various teams around the world have developed methods for comprehensive environmental DNA analysis of fish, but the method using the MiFish primer is now becoming the standard. This method was developed by Dr. Masaki Miya of the Natural History Museum and Institute, Chiba, and we were also involved in its development. Several papers have compared methods using MiFish primers with methods using primers made by teams from other countries, such as Europe or the US. These papers have concluded that the detection methods using MiFish primers are the best or better than the others. The "Environmental DNA Sampling and Experiment Manual" also describes the methods using MiFish primers.
The revised English edition was released in April 2019 (https://ednasociety.org/wp/wp-content/uploads/2020/09/eDNA_manual_Eng_v2_1_3b.pdf). For the revision, we solicited opinions on the manual and received numerous requests. There were many requests to include details for portions we had thought were not very important, portions where we had decided it would be better to be flexible, and portions that were not yet set in stone. The revised edition does not cover every aspect; however, after the next edition, I would like to include the opinions we weren’t able to this time. We plan to make minor updates every year if possible, and major updates every two or three years. The video related to the manual will also be released by the society. As the number of researchers and research subjects increases in the future, work like making revisions will become more difficult, but I think it is necessary to popularize the methods.
Q. Which part of "Environmental DNA Sampling and Experiment Manual" is the most important?
A. The comprehensive explanation on how to prevent contamination in the laboratory.
Contamination is our worst enemy when we try to detect very diluted DNA from only a few copies in the sample. Before the manual was created, there were queries saying, "We performed the analysis exactly as written in the paper, but it didn't work." There is no detailed description of the know-how in the paper, so we have provided information for those who sent us direct inquiries. The manual explains that conventional methods, particularly in laboratories, are prone to introduce “carry-over contamination,” where PCR products get mixed into samples. However, even in our own laboratory, we still have trouble eliminating contamination, which is why we need to take measures from a variety of aspects, such as improving the environment, creating rules, and devising data processing techniques.
Q. What are the difficulties in analyzing the data generated by NGS?
A. In some cases, DNA sequences alone cannot distinguish one species from close relatives.
In some cases, the species cannot be determined based on the sequences generated by the NGS analysis. For example, the sequence of black sea bream in the genetic region amplified using MiFish primers is the same as that of Okinawa seabream inhabiting the south. The sequence we examined in Maizuru Bay is the one believed to be from black sea bream, but strictly speaking, it cannot be distinguished from Okinawa seabream. By knowing that Okinawa seabream should not be in Maizuru Bay, we determined that they were black sea bream. This sort of speculative step may have to be included mid-process. For more reliable species identification, you should also test different genetic marker areas from that of MiFish, but this will require more effort and may also compromise the quality of the environmental DNA analysis.
Another problem is that, in a surprising number of cases, the species name registered in the genetic database is found to be incorrect. The international genetic database we use is self-registered by all researchers around the world. The agencies in charge in each country conduct a simple review, but they don't check details of contents, so potentially wrong information cannot be checked. It's not easy to ask the registrant to correct it, and there is no countermeasure against it. For this reason, the Ministry of the Environment is currently trying to create a robust database by scrutinizing the data. I think that this kind of work will have to be taken over by the society someday.
For Japanese freshwater fish, more than 90% of the sequences are listed in the database, so most of them can be identified, except for very minor ones. For saltwater fish along the coast of Japan, it is estimated that 30 to 40% of them can be identified. For that reason, it is important to design a database and specific primers.
Q. How did environmental DNA analysis and your own research begin?
A. They began in the 2000s with environmental DNA analysis for viruses.
In the area of environmental DNA analysis, the analysis of microorganisms became popular from the 1990s to the early 2000s.
The first study of visible-sized organisms (or macroorganisms) was conducted in 2008 by a French team and they reported on the distribution of bullfrogs by analyzing pond water. Since then, research on species-specific environmental DNA has progressed simultaneously in several places around the world, including a survey of exotic species of carp in the Great Lakes in the United States.
My own environmental DNA analysis of viruses started around 2007. By investigating the relationship between the carp-infecting carp herpesvirus and carp and human life at the Research Institute for Humanity and Nature (RIHN), Kyoto, I discovered seasonal variations and regional differences of the carp herpesvirus in Lake Biwa. Then, I came up with the idea that the amount of host carp and the state of the infection could be estimated from the amount of virus released from a single carp, and conducted a preliminary study. The result found that the total amount of DNA in the water was very large, and the carps themselves released DNA in various forms, such as feces and mucus. When I was sharing these findings with Dr. Hiroki Yamanaka (Advanced Science and Technology, Ryukoku University), a collaborator and fish ecologist, it occurred to me that it might be possible to understand the distribution of fish by studying water samples, and since then I have been working on the research. The first successful example of detecting environmental DNA from wild fish was the detection of DNA from Acheilognathus rhombeus, a member of bitterlings, in water from Lake Ibanai (Higashiomi City, Shiga Prefecture, Japan), one of the inner lakes of Lake Biwa.
Since 2009, we have been working on what we now call environmental DNA metabarcoding. We steadily performed DNA sequencing using the Sanger method after random cloning of the DNA from five species of fish in a water tank. At that time, we weren’t doing NGS analysis, so the DNA sequencing was very hard. The paper of this study was published electronically in 2011 and is the first paper on comprehensive environmental DNA analysis of macroorganisms (Surveillance of fish species composition using environmental DNA, Toshifumi Minamoto, Hiroki Yamanaka, et al., Limnology volume 13, pages 193 - 197 (2012), https://link.springer.com/article/10.1007/s10201-011-0362-4).
Q. Please let us know about your recent research.
A. We are conducting research by combining species-specific environmental DNA analysis with comprehensive environmental DNA analysis.
We studied a zoonosis called leptospirosis.1 Leptospires are carried in the kidneys of wild animals, domestic animals and pets, and are excreted in urine. Humans can be infected directly through this urine, or through water or soil containing the urine. Symptoms range from a mild cold, to in severe cases, jaundice and kidney damage. The number of patients in Japan is not large, but in Okinawa Prefecture in particular, cases of infection during leisure activities on the coast were found, and the host was unknown. So, we sampled water from two rivers in Okinawa Prefecture, and repeatedly performed a specific analysis of leptospires in the water, as well as a comprehensive analysis of vertebrates. As a result, we found that wild boar or swine DNA came out together with leptospira DNA. It can be presumed that wild boars carry leptospires. Results like this can be obtained when a specific analysis of infectious agents and a comprehensive analysis of natural host candidates are performed simultaneously.
There is also some debate as to when specific environmental DNA can be properly detected in water. In summer, it is assumed that more DNA is released because the activity of living organisms increases. On the other hand, the decomposition of released DNA is also enhanced because the water temperature is high and the microbial activity increases. On the contrary, in winter, it is generally assumed that less DNA is released and that degradation is slower. So, you can't tell which season is the best until you investigate. We sampled water from three dam lakes throughout the year and this revealed that the highest number of fish species can be detected in spring.2 This is because many species lay eggs in spring, individuals are active during the laying period, sperm and eggs are released, and young fish have high metabolic properties, all of which results in increased environmental DNA.
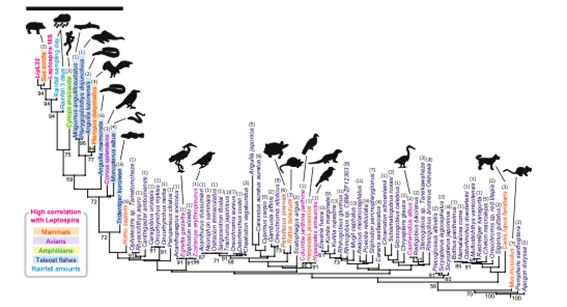
Figure 1. Positive correlation between the detection pattern of leptospiral DNA and that of wild boar/swine genes or rainfall pattern was found.
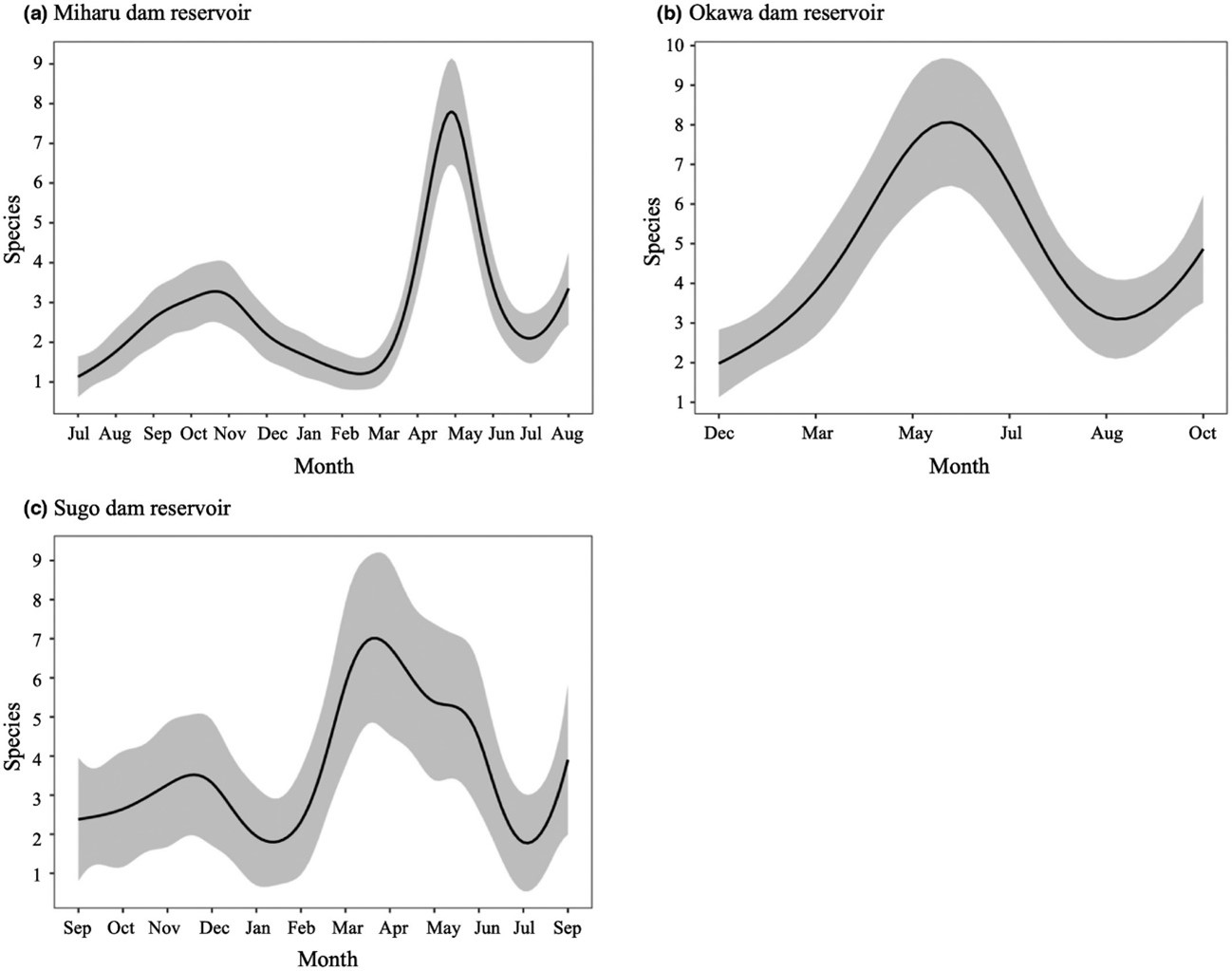
Figure 2. A comprehensive analysis of fish environmental DNA was carried out using water samples collected from Miharu Dam and Okawa Dam in Fukushima Prefecture and Sugo Dam in Hyogo Prefecture from 2015 to 2016. It revealed that the number of fish species that can be detected began to increase in all three dams after March and peaked around April and June.
Q. What kind of research are you planning to do in the future?
A. I would like to do research that will advance environmental DNA analysis.
As basic research, I would like to capture physiological conditions, such as stress and reproductive behavior of living things in the environment. If it is difficult to capture it using DNA, it may be possible to capture it using RNA.
As an applied research, we are considering whether information on the distribution of fish can be used for fisheries, or whether information on the distribution of parasites and pathogens can be used for the health of humans and other animals.
I also feel the potential for research on restoration of past biological information. For sardines and horse mackerels in the ocean, research is being carried out to compare the amount of DNA in sediment with the volume of landings. Similarly, if you core the sediment at the bottom of the lake and perform a comprehensive analysis from top to bottom, you may find the transition of the fish fauna in that location. So far, we've found that we are likely able to capture at least 300 years of change. If we can also incorporate environmental information, such as water, temperature and water quality, we may be able to estimate the impact of human activities.
The interesting thing about environmental DNA analysis is that once you've taken a sample, you can reuse it later for various purposes. We can see a new world by consolidating biological information from microorganisms to macroorganisms using environmental DNA analysis.

Field research in Okayama Prefecture. "The fun part of this research is that we can go to many places in search of water".
References:
- Environmental DNA metabarcoding to detect pathogenic Leptospira and associated organisms in leptospirosis-endemic areas of Japan
Y Sato, et al., Scientific Reports, (2019) 9:6575 https://doi.org/10.1038/s41598-019-42978-1 Hayami K, Sakata MK, Inagawa T, et al. - Effects of sampling seasons and locations on fish environmental DNA metabarcoding in dam
Ecol Evol. 2020;10:5354–5367. https://doi.org/10.1002/ece3.6279
Learn more about the products and systems mentioned in this case study:
Illumina NovaSeq 6000 System
www.illumina.com/novaseq
Environmental DNA Analysis page
https://sapac.illumina.com/destination/edna.html
