がん研究アプリケーション
リキッドバイオプシー研究、シングルセル解析、エピジェネティクスなど、がん研究におけるさまざまなアプリケーションを探ります。
腫瘍の免疫原性と免疫療法の反応性に関する理解を深める
がん免疫研究を牽引する研究者らは、次世代シーケンサー(NGS)を活用し、免疫療法反応因子、バイオマーカー、ゲノムなどを研究することで、個別化免疫療法の研究を向上させています。
腫瘍が免疫反応を回避するために用いるメカニズムに関するがん免疫研究は、有望な治療ターゲットにつながっています。これらの療法により、免疫システムのがんを標的とする能力が高まったり、腫瘍が免疫反応を回避する能力が抑制されたりします。さらに、腫瘍環境においてどのパスウェイが活性化され、それががん細胞の増殖、生存、浸潤、転移などのプロセスにどのように関わっているかを特定するうえで、NGSはきわめて有用となります。

がん細胞が免疫による検出を回避するために用いる複雑な経路に関する理解は、急速に深まり続けています。主要な研究者らは、NGS技術を活用してバイオマーカーを発見し、個別化された免疫療法の将来の開発の指針となる分析ツールを開発しています。このアプローチで重要なのは、持続的な抗腫瘍免疫反応を媒介できる適切なネオアンチゲン反応性T細胞を同定することです。1
NGSは、がんゲノムの詳細な解析を提供できる強力なツールです。また、腫瘍増殖または治療に反応する免疫マーカー発現をモニタリングするために、高感度で腫瘍微小環境をリアルタイムで効率的に評価することも可能です。NGSは、免疫細胞のレパートリーを特徴付け、微小環境内のさまざまな細胞集団を同定し、同時に数千のターゲットにおける遺伝子発現を包括的に定量することができます。
NGSは、研究者らがネオアンチゲンを同定し、免疫反応を高める革新的な治療法を研究し、遺伝的変異がその効果に与える影響を解明するうえでも役立ちます。さらに、NGSは、腫瘍特異的な反応を誘発するために適用できる新規の腫瘍抗原の予測的な選択も可能にします。
空間トランスクリプトミクスにより、組織切片にマッピングされた遺伝子発現パターンのトポグラフィー配置が得られるほか、構造と活性を関連付けることが可能です。この能力により、研究者は細胞レベルで生物学的相互作用を明確にし、腫瘍微小環境などの複雑な組織に対する新しい洞察を得ることができます。
宿主微生物相互作用ががんの発生、進行、治療効果に及ぼす影響を研究するために、NGSを用いることができます。NGSにより、さまざまな状況で微生物群をプロファイルできるため、このプロファイリングは新たながん治療法の開発でターゲットとなりうる種や症状を同定するうえできわめて重要です。
シングルセルRNAシーケンス(scRNA-Seq)は、シングルセル解像度で、がんや腫瘍微小環境のトランスクリプトームプロファイルを調べるためにますます利用されるようになっています。免疫細胞研究は、シングルセルトランスクリプトミクスと組み合わせて、がんの免疫サーベイランスと免疫療法に対する耐性における主要な因子を理解するために適用されてきました。
免疫レパートリーシーケンス(IR-Seq)では、NGSを使用してB細胞およびT細胞受容体レパートリーをプロファイルします。IR-Seqにより、サイエンティストはさまざまながんにおけるリンパ球受容体バリアントの研究、血液悪性腫瘍研究などが可能となります。
このアプリケーションノートでは、進化する傾向、研究者のニーズ、この急速に進化する分野で利用できるゲノムテクノロジーなど、免疫腫瘍学の最近の進歩を強調しています。
NGS法により、免疫チェックポイント発現、腫瘍変異負荷(TMB)、腫瘍微小環境、腸内微生物組成における重要な変化を高い分析感度で研究することが可能になります。
チェックマーク
|
主なアプリケーション |
| ネオアンチゲン、変異負荷 | 発現プロファイリング | マイクロバイオーム(16S)シーケンス | TCR/BCRaプロファイリング | エピジェネティックなプロファイリング | |
|---|---|---|---|---|---|
| 治療への応用 | |||||
| チェックポイント阻害剤 | チェックマーク
|
チェックマーク
|
チェックマーク
|
チェックマーク
|
|
| ワクチン | チェックマーク
|
チェックマーク
|
チェックマーク
|
チェックマーク
|
|
| 養子細胞療法 | チェックマーク
|
チェックマーク
|
チェックマーク
|
チェックマーク
|
|
| 予後 | |||||
| 微生物 | チェックマーク
|
||||
| 免疫レパートリー | チェックマーク
|
チェックマーク
|
チェックマーク
|
||
| がんモニタリング | チェックマーク
|
チェックマーク
|
チェックマーク
|
リキッドバイオプシー研究、シングルセル解析、エピジェネティクスなど、がん研究におけるさまざまなアプリケーションを探ります。
単一遺伝子およびアレイベースのアプローチよりも短時間でより多くの情報を提供する次世代シーケンサー(NGS)ベースのがんシーケンス法について学びましょう。

がん研究手法に関するガイドは、幅広いがん研究アプリケーションのためのシンプルで包括的なワークフローを紹介した40ページ以上にわたる包括的なリソースです。このガイドには、シングルセルシーケンス、空間シーケンス、メチル化プロファイリング、マルチオミクス、セルフリーRNAシーケンスなどが含まれます。
ご提供いただいた個人情報は、お客様へのサポート、サービス、および販売活動の目的にのみ使用させていただきます。
マルチオミクス研究は、ゲノム、エピゲノム、トランスクリプトーム、メタゲノム、プロテオミクスのアプローチからの高次元データセットを統合し、多くの場合、計算およびネットワーク生物学を使用して、これらの技術によって生成される膨大な量のデータを解釈します。臨床研究データにマルチオミクスを適用することにより、既存のオミクス手法の探索力が強化され、新しいバイオマーカーや免疫療法のターゲットが明らかになります。
さらに、マルチオミクスアプローチは、腫瘍および微小環境の分子プロファイルの包括的な見方を提供することができます。複数の情報レイヤーにより、単一オミクス研究だけでは解決できない腫瘍生物学に対する新たな洞察が得られます。

Illuminaでは、がん免疫研究に対応したデータ解析オプションが利用できるライブラリー調製およびシーケンスオプションを複数ご用意しています。効率化されたワークフローとフレキシブルなキット構成で、さまざまな研究デザインに対応できます。

AIとインフォマティクスにおけるイノベーションによって、どのようにNGSの複雑さが簡素化され、幅広いラボで臨床研究ができるようになるのかをご覧ください。
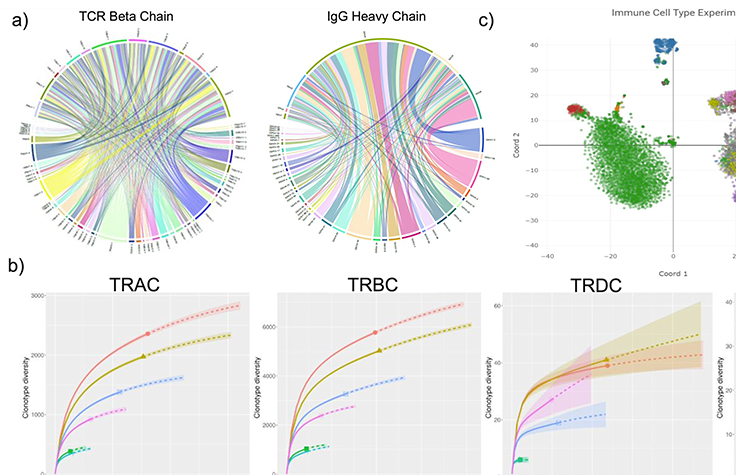
このダウンロード可能なポスターは、完全長V(D)J免疫レパートリーシーケンス(IR-Seq)を可能にする、がん研究用の新しいスケーラブルなツールに関する概要を示しています。

弊社のシーケンステクノロジーとマイクロアレイテクノロジーは、DNAからRNA解析、エピジェネティクスなど、がんゲノム研究の応用分野を幅広くサポートしています。
がん免疫研究メソッドについて、より詳しい情報をご希望の場合は、お気軽にお問い合わせください。
ご提供いただいた個人情報は、お客様へのサポート、サービス、および販売活動の目的にのみ使用させていただきます。