コロナウイルスのシーケンス
NGSがCOVID-19対策に役立つ理由
次世代シーケンサー(NGS)は、微生物に関する事前知識がなくても、新しいコロナウイルス株やその他の病原体を効果的かつ偏りなく同定できます1。シーケンスは、感染拡大の初期にCOVID-19(SARS-CoV-2)の原因となる新型コロナウイルスを同定するために使用されました2。SARS-CoV-2コロナウイルスの急速に広がる新しい変異株への懸念が続いており、変異を迅速に検出して新株の拡散を防ぐために、シーケンスの実施を強化する必要性が高まっています。NGSは、公衆衛生当局、ワクチンや薬剤の開発者、研究者に重要なエビデンスを提供し、それによりラボは以下のことが可能になります:
- ウイルスの感染経路をグローバルに追跡
- 変異を迅速に検出し、新変異株の伝播を防止
- ワクチンの効能に影響を与える、あるいは確立された分子診断法による検出を回避しうるウイルスの変異を同定
- COVID-19の治療薬候補となる標的のスクリーニング
- 呼吸器複数感染および薬剤耐性アリルの同定と特性解析
SARS-CoV-2の新規株を同定・追跡するためのソリューション
動画プレゼンテーションで、イルミナが全従業員の献身と協力を得て、記録的な速さでCOVIDSeq Testを開発した様子をご覧ください。さらに、COVIDSeq Assayにより、SARS-CoV-2の新規株の出現と有病率をラボがどのように特定・追跡できるかについても紹介しています。
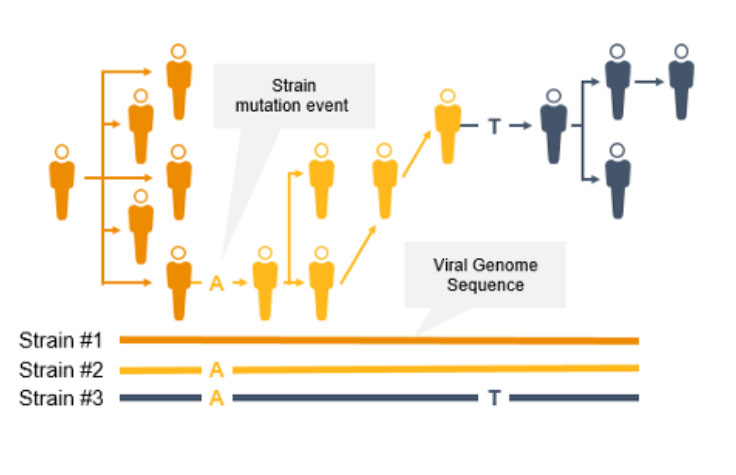
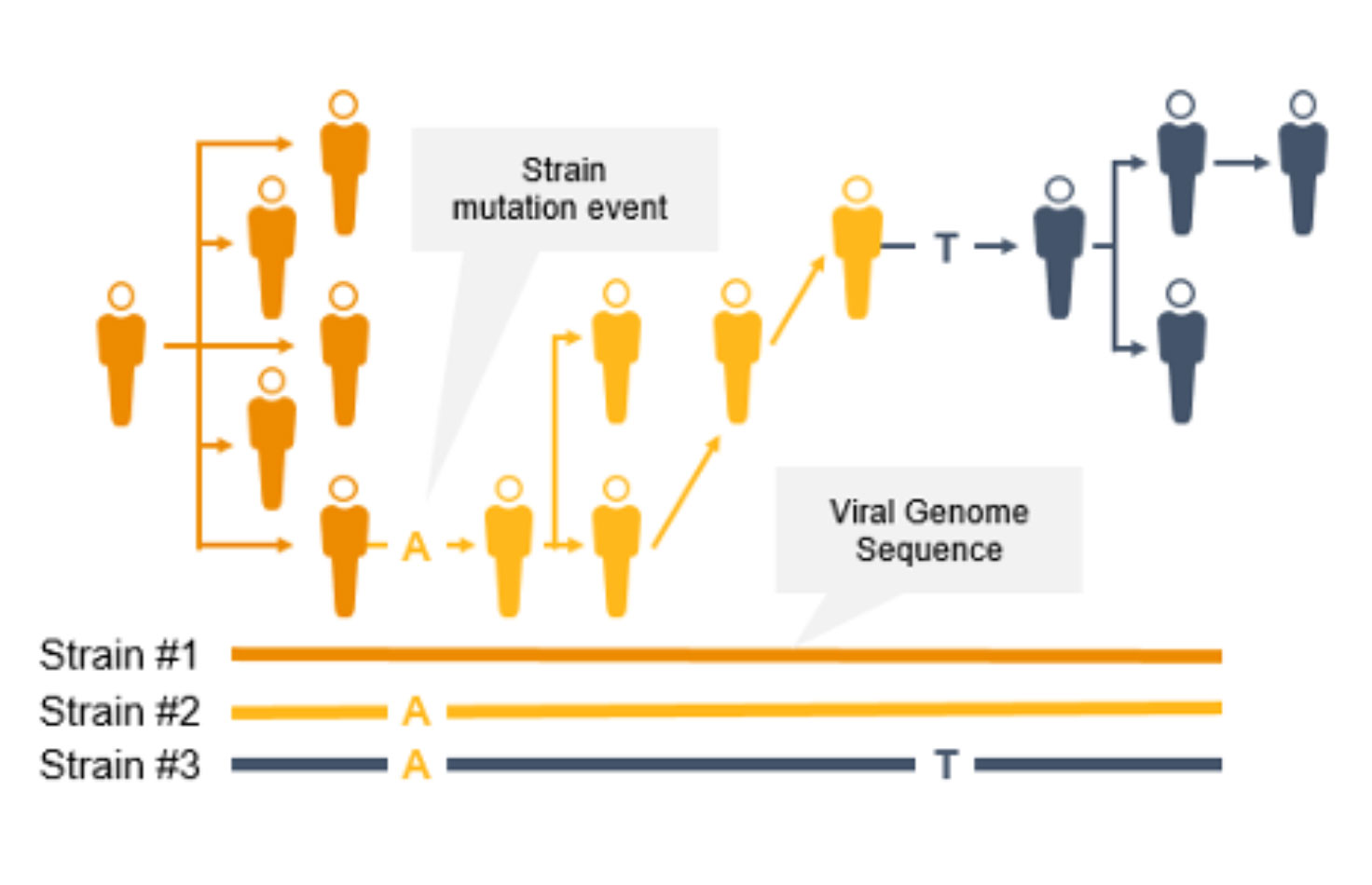
NGSによるコロナウイルス変異の検出と特徴づけ
コロナウイルスは変異するため、新たな変異株が出現し、一般の人々に影響を及ぼす可能性があります。突然変異によって伝播性や感染性が増大する可能性があるため、サーベイランスとモニタリングが不可欠になります。このようなコロナウイルスの変異は、ワクチンの効果/保護を低下させたり、検査によって診断されにくくなったりする可能性があります3。
NGSは、COVID-19などの感染症のゲノムサーベイランスのための貴重なテクノロジーです。NGSは、コロナウイルス変異株の蔓延を追跡できるだけでなく、PCRとは異なり、新型コロナウイルス変異を同定することもできます。NGSを使用して、サイエンティストは低頻度のマイノリティバリアントと複数の多型および新型バリアントを検出できます。
NGSを使用したゲノムワイドな系統タイピングは、疫学者が突然変異を迅速に同定して特徴づけることで、感染拡大防止に役立ちます。系統レベルの追跡によって、感染拡大クラスターと伝播経路の特定をサポートできます。
対照的に、PCRは病原体ゲノムの特定の領域を検出するように設計されているため、急速に進化する病原体ゲノム全体で新たな突然変異を同定することはできません。さらに、プライマーまたはプローブの結合領域に変異が生じると、PCRの性能が低下する可能性があります。
コロナウイルスシーケンス法の概要
アンプリコンシーケンス
全ウイルスゲノムのシーケンスによりSARS-CoV-2コロナウイルスを検出し、特徴づけます。この方法では、PCRアンプリコンのウルトラディープシーケンスにより、ゲノム全体を解析できます。
ハイブリッドキャプチャーシーケンス
コロナウイルス、インフルエンザウイルス、その他の呼吸器病原体、および関連する薬剤耐性アリルを検出し、その特性を解析します。こうした知見は、研究者が呼吸器感染症をモニタリングし、感染制御戦略を最適化する際に役立ちます。この方法により、ターゲット特異的なプローブとのハイブリダイゼーションを介して、所期のゲノム領域を捕捉することができます。
ショットガンメタゲノミクス
特定のサンプルに含まれるすべての微生物を包括的にシーケンスし、コロナウイルスなどの新しい病原体を同定することができます。このNGS法は、感染拡大調査を迅速化し、新しいラボ検査の開発を支援する上で、役立つ可能性があります。
コロナウイルスNGS法の比較
| SARS-CoV-2のサーベイランス | 呼吸器病原体の検出とサーベイランス | 新型ウイルスの検出 | |
|---|---|---|---|
| 検出に必要なもの | アンプリコン | ハイブリッドキャプチャー | ショットガンメタゲノミクス |
| スピードとターンアラウンドタイム | |||
| スケーラビリティと費用対効果 | |||
| 新規病原体の同定 | |||
| 伝播の追跡 | |||
| 変異の検出 | |||
| 複数感染や複雑な疾患の同定 | |||
| 薬剤耐性の検出 |
ラボの検査ニーズを十分に満たします
ラボの検査ニーズを部分的に満たします
アンプリコンシーケンス、ハイブリッドキャプチャー、メタゲノムワークフロー
一緒に購入されることの多い製品
コロナウイルスソフトウェアツール
これらのツールは、コロナウイルスのシーケンスデータ解析を加速し、サンプル追跡を簡略化し、宿主応答研究を促進するものです。
DRAGEN二次解析
ウイルスや微生物のシーケンスからRNA-Seq、全ゲノムシーケンスまで、幅広いアプリケーションに対応する強力で最先端のNGSデータ解析ツールを利用できます。
Correlation Engine
このインタラクティブなオミクスの知識ベースとデータ検索エンジンは、プライベートデータを、キュレーションされた公開オミクスデータを用いて生物学的なコンテキストに取り込み、研究者がCOVID-19の宿主応答を制御する要因を調査するのに役立ちます。
Clarity LIMSソフトウェア
この高度にカスタマイズ可能なラボ情報管理システムにより、ラボはサンプルを追跡し、ワークフローを効率的かつ安全に管理することができます。
COVID-19 NGSウェビナー
SARS-CoV-2対策におけるゲノムの応用
感染症研究の第一人者である2人の研究者が、ゲノムシーケンス技術によるCOVID-19のパンデミックへの取り組みについて語ります。
SARS-CoV-2に対する臨床メタゲノム解析
COVID-19パンデミックにより、SARS-CoV-2のような新興病原体を検出および監視するツールの必要性が浮き彫りになっています。NGSはコロナウイルスの早期検出につながり、検査やワクチン開発を加速します。
SARS-CoV-2検出と監視のためのNGS
新型コロナウイルスの早期検出と特性解析から、モニタリング、サーベイランス、診断検出まで、COVID-19パンデミックへの対応におけるNGSの幅広い適用性についてご紹介します。
呼吸器系ウイルスシーケンスアプリケーションノート
呼吸器病原体と薬剤耐性の検出
呼吸器病原体(SARS-CoV-2、インフルエンザウイルス、真菌を含む)と関連する薬剤耐性アリルを幅広く検出するための迅速ハイブリッドキャプチャーシーケンス。
Respiratory Pathogen ID/AMRワークフローアプリケーションノートRespiratory Pathogen ID/AMRワークフローの分析性能アプリケーションノート
呼吸器系ウイルスを発見して特徴付ける
コロナウイルス、インフルエンザウイルス、その他の呼吸器系ウイルスの高感度検出と特性解析のための迅速ハイブリッドキャプチャーシーケンスワークフロー。
呼吸器系ウイルスの迅速な検出
MiniSeq Rapid試薬は、シーケンスランタイムを5時間未満に短縮し、コロナウイルスやその他の呼吸器系ウイルスの迅速な検出を可能にします。
最先端の臨床研究用シーケンスラボを設立
Shamir Genome CenterのDr. Nir Rainyが、ハイスループットのゲノミクスセンターの構築に何が必要か、イスラエルにおけるCOVID-19サーベイランスの取り組みをどのように支援しているか、臨床研究におけるNGSの将来に対するビジョンについて述べています。
記事はこちらCOVIDシーケンスの顧客事例
COVID-19シーケンスと共に道を切り拓く
ユタ州公衆衛生研究所のディレクターが、SARS-CoV-2感染拡大クラスターに狙いを定め、将来的なパンデミックに向けたインフラを確立するための取り組みにおいて、どのようにNGS業務を拡大したのかについて説明します。
オーストラリアにおけるCOVID-19追跡システムの構築
画期的な取り組みとして、公衆衛生研究所はイルミナの技術を用いて、オーストラリアにおける全ての陽性COVID-19検査のウイルスゲノムのシーケンスを行い、州ごとではなく、国全体でCOVID-19を追跡しています。
COVID-19宿主応答研究を促進
共同作業の環境により、COVID-19の研究者は、重要な経路やバイオマーカー、医薬品候補となるリード化合物に関する仮説を検証できます。
変化するパンデミック下でのCOVIDシーケンスの改善
Aegis Sciencesは、いかに変異に対応し、能力を拡大し、世界最大のCOVIDシーケンス事業の1つになったか。
オハイオ州立大学での粉塵によるCOVID-19の追跡
キャンパスラボは、SARS-CoV-2やその他のウイルスのバリアントとウイルス量を理解するために、真空バッグから粉塵をシーケンスします。
技術的なヒント
新型コロナウイルスに対するイルミナシーケンサーの汚染除去について
米国CDCおよび世界保健機関の勧告に従い、イルミナは新型コロナウイルスSARS-CoV-2(2019-nCoV)に接触した疑いがある、または接触が判明しているNGS機器関連の汚染を除去するためにこの手順を推奨しています。
アンモニウム系洗浄剤によるシーケンスラン性能への影響
アンモニア系洗浄剤および消毒剤(COVID-19パンデミック時にラボの洗浄に頻繁に使用されているもの)をシーケンスランの設置場所の近くで使用すると、シーケンスランの性能が低下する可能性があります。これらの問題を避けるためのヒントを見る。
あなたのラボのCOVID-19シーケンスソリューションについてご相談ください。
以下お問い合わせください
関連ソリューション
結核サーベイランス
ゲノムベースの結核サーベイランスが、公衆衛生当局による感染拡大の検出や伝播の追跡にどのように役立つかをご覧ください。
抗菌薬耐性サーベイランス
さまざまなNGS法が、抗菌薬耐性の検出にどのように革新的な能力を提供できるかについて知見が得られます。
廃水シーケンス
NGSによる廃水サーベイランスが、新興感染症のコミュニティーレベルの監視、変異の追跡、変異株の動向把握にどのように役立つかを学べます。
参考文献
- Bulcha B. Review on viral metagenomics and its future perspective in zoonotic and arboviral disease surveillance. J Biol Agr Healthc. 2017; 7(21): 35–41.
- Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020; 382(8): 727–733.
- Tracking SARS-CoV-2 variants. who.int/activities/tracking-SARS-CoV-2-variants. アクセス日:2023年12月6日。